 |
| Carl Woese in 2006, via U Illinois |
For this insight, we can thank Carl Woese, who passed away just before the New Year at the age of 84 (U Illinois press release, NY Times obituary). Woese made his discovery in the 1970s, when sequencing DNA and RNA was anything but routine. At that time, the prokaryotes were classified primarily by physiological characteristics and morphological appearance. There was little attention paid to microbial evolution, except in a speculative way, simply because there was no easy way to go about studying it.
Woese's insight was to recognize that ribosomal RNA could be used as a marker of evolution. Every living cell contains ribosomes, and both their structure and function are conserved, meaning that even distantly-related organisms could be compared using their rRNA. But rRNA sequences are not completely frozen in time: they evolve at a rate that's well-suited to reconstructing the evolutionary relationships between the major groups of life.
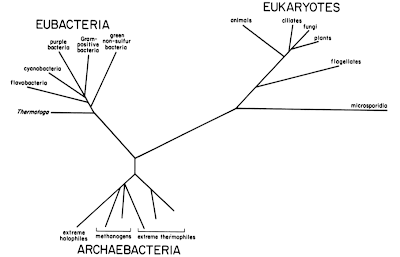 |
| Ribosomal tree of life, showing the three domains, from Woese 1987. |
Today, sequencing rRNA is routine work: we use PCR to amplify the sequences, and then sequence them using one of a variety of technologies. In Woese's day, this was not possible. What he used instead was a method called "oligonucleotide cataloguing". First the rRNA had to be physically isolated from the cells of an organism. They were then cut into fragments at every G residue by an enzyme derived from fungi, called ribonuclease T1. These fragments were then separated by electrophoresis on
It was with oligonucleotide cataloging that he discovered the third domain of life, when he compared the catalogs from methanogens, a group of prokaryotes that produce methane as a by-product of their metabolism, to other organisms. He found that they were as distant from the bacteria as they were from the eukaryotes: despite looking to other microbiologists like bacteria, they were instead a totally new group of life! Other lines of evidence apart from rRNA have since confirmed the uniqueness of the Archaea, but when his results first came out, few people believed him or thought that it was important. Lynn Margulis, for example, still favored the term "prokaryote" because in her system, eukaryotes represented a wholly new grade of cellular organization over the prokaryotes. The comments by Joe Felsenstein in this post on the Panda's Thumb blog provide a good idea of the context in which Woese was proposing his new ideas.
It took a long time for Woese's views to win out over his detractors, a battle which reportedly left him a "scarred revolutionary", according to a 1997 profile in Science (also available here). Some of his skeptics could afford to hold out well into the 1990s, because they were already established names (such as Ernst Mayr). Today, the use of rRNA as a phylogenetic marker is pervasive. The small-subunit rRNA gene is the most widely-used phylogenetic marker, and there exist specialized databases devoted to rRNA data, such as CRW and SILVA. The three domains are textbook dogma, but current research is moving beyond using single molecules to reconstruct phylogeny. Using multiple genes is now the standard for serious phylogenetics, and phylogeny is being used as a guide for genome sequencing projects.
Microbiology was one of the last major fields in biology to thoroughly incorporate an evolutionary perspective, and Carl Woese was the individual who did the most to cause that shift. Indeed it may have even gone a bit too far: because sequencing is much easier than isolation and cultivation of new organisms, we now have a surfeit of genomic sequences from environmental organisms, but a limited set of cultivated microbes for performing experiments that help us make sense of these genomes. The future for environmental microbiology appears to lie in making the best use of new high-throughput sequencing technologies with targeted cultivation of selected strains, to both broaden and deepen our understanding.
1 comment:
Separation of T1 oligos was by PAPER electrophoresis (Cellulous acetate and DEAE-81) not by gels.
Post a Comment